Systems Medicine
Research Lead
General aim
The discovery and development of new and potentially more potent drugs suffers from a conceptual crisis affecting medicine in general: Diseases are currently defined and diagnosed based on symptoms (e.g., hypertension, depression) or affected organs (e.g., heart failure, nephropathy). There is a lack of mechanistic understanding of diseases that could improve disease definitions and allow for more informed selection and development of therapies.
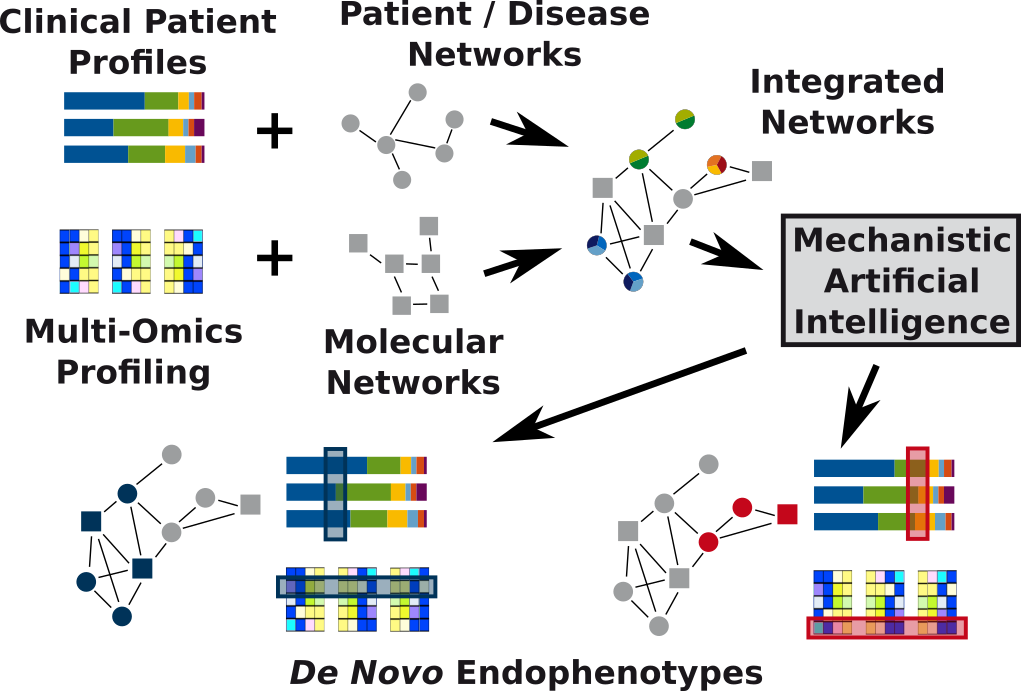
The novel field of systems medicine addresses these issues. Among important research goals are mechanistic definitions of diseases, drug repurposing, and de novo endophenotyping. Systems medicine is highly inter-disciplinary and can only strive when biologists, clinicians, and computer scientists work closely together.
Focus of research
Modern omics technologies rapidly become cheaper, more exhaustive, and more precise. Furthermore, there is an immense growth in public databases on molecular interactions, comorbidities, drug effects, etc. Integrative methods can leverage this situation to change future clinical decision-making. In particular, network-biology and machine learning are central pillars of the necessary methodology complementing multi-omics studies. This becomes evident in de novo endophenotyping, which encompasses the stratification of patients based not only on simple molecular markers, but on composite network-based markers. Big data analytics will allow for in silico trials, thus speeding up drug repurposing and the development of novel (composite) biomarkers and therapeutic approaches. The success of systems medicine hinges on the availability of large data sets. Thus one important technological and societal question is how we can break down barriers that prevent data sharing across medical institutions.





